shinyButchR: Decompose Data with Non-negative Matrix Factorization
★ ★ ★ ★ ☆ | 1 reviews | 3 users
About the app
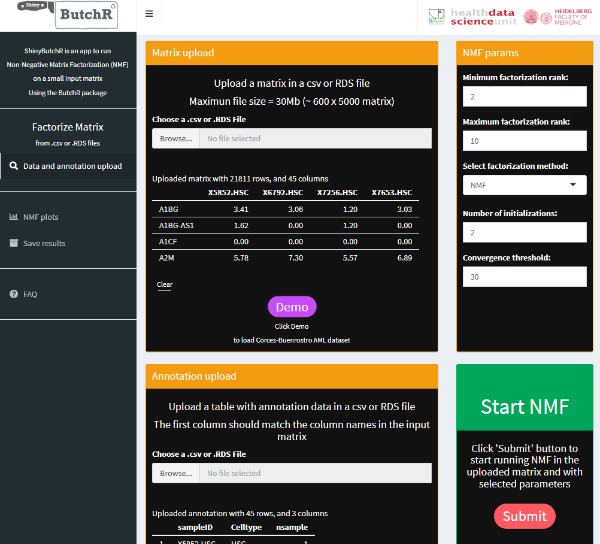
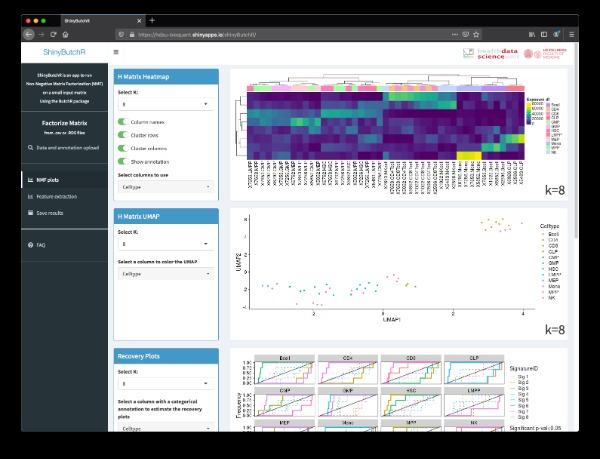
ShinyButchR is an interactive R/Shiny app to perform NMF on an input matrix.Non-negative matrix factorization (NMF) has been widely used for the analysis of genomic data to perform feature extraction and signature identification due to the interpretability of the decomposed signatures. ShinyButchR uses {ButchR} R package to run the matrix decomposition and generate diagnostic plots and visualizations that helps to find association of the NMF signatures to known biological factors. Original publication: Andres Quintero, Daniel Hübschmann, Nils Kurzawa, Sebastian Steinhauser, Philipp Rentzsch, Stephen Krämer, Carolin Andresen, Jeongbin Park, Roland Eils, Matthias Schlesner, Carl Herrmann, ShinyButchR: interactive NMF-based decomposition workflow of genome-scale datasets, Biology Methods and Protocols, bpaa022.
Data Safety
Safety starts with understanding how developers collect and share your data. Data privacy and security practices may vary based on your use, region, and age. The developer provided this information and may update it over time.
Rate this App
App Updates and Comments
No application notes or reviews.
Post response or reviewsOther Similar Apps
About ShinyAppStore
ShinyAppStore is the leading platform for showcasing and utilizing a wide range of Shiny applications developed with the shiny R package. We will be expanding access to allow shiny apps built with Python as well. Users can submit and explore applications across various categories, add verified apps to their personal library, and enjoy easy access. The platform features well-designed apps with detailed descriptions and evaluations by users. All applications on ShinyAppStore are user-owned and open source, with the source code readily available for download on our GitHub page.
More "about" details